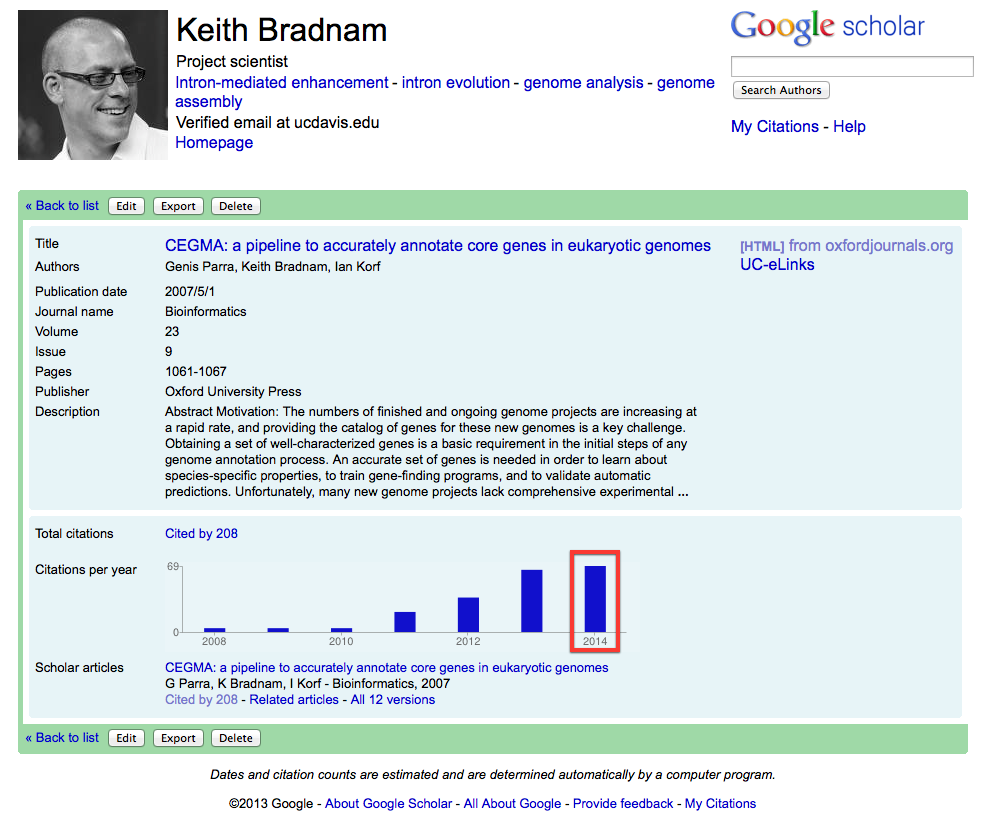
This was because it had fallen on me to continue to deal with all CEGMA-related support requests. Until 2010, there hadn't really been any support requests because almost no-one was using CEGMA. This changed dramatically and I started to receive lots of emails that:
- Asked questions about interpreting CEGMA output
- Reported bugs
- Asked for help installing CEGMA
- Suggested new features
- Asked me to run CEGMA for them
I started receiving lots of the latter requests because CEGMA is admittedly a bit of a pig to install (on non Mac-based Unix systems at least). In the last 6 months alone, I've run CEGMA 80 times for various researchers who (presumably) are unable to install it themselves.
After the version 2.3 release — necessary to transition to the use of NCBI BLAST+ instead of WU-BLAST — and 2.4 release — necessary to fix the bugs I introduced in v2.3! — I swore an oath never to update CEGMA again. This was mostly because we no longer have any money to work on the current version of CEGMA. However, it was also because it is not much fun to spend your days working on code that you barely understand.
It should be said that we do have plans for a completely new version of CEGMA that will — subject to our grant proposal being successful — be redeveloped from the ground up, and will include many completely new features. Perhaps most importantly — for me at least — a version 3.0 release of CEGMA will be much more maintainable.
And now we get to the main source of my ire when dealing with CEGMA. It is built on a complex web of Perl scripts and modules, which make various system calls to run BLAST, genewise, geneid, and hmmsearch (from HMMER). I still find the scripts difficult to understand — I didn't write any of the original code — and therefore I find it almost impossible to maintain. One of the reasons I had to make this v2.5 update is because the latest versions of Perl have deprecated a particular feature causing CEGMA to break for some people.
Most fundamentally, the biggest problem with CEGMA (v2.x) is that it is centered around use of the KOGs database, a resource that is now over a decade old. This wasn't an issue when we were developing the software in 2005, but it is an issue now. Our plans for CEGMA v3.0 will address this by moving to a much more modern source of orthologous group information.
In making this final update to v2.x of CEGMA, I've tried adopting some changes to bring us up to date with the modern age. Although the code remains available from our lab's website, I've also pushed the code to GitHub (which wasn't in existence when we started developing CEGMA!). In doing this, I've also taken the step to give our repository a DOI and therefore make the latest version citable in its own right. This is done through use of Zenodo.
Although I hope that this is the last thing that I ever have to write about CEGMA v2.x, it is worth reflecting on some of the ways that the process of managing and maintaining CEGMA could have been made easier:
- Maintain documentation for your code that is more than just an installation guide and a set of embedded comments. From time to time, I've had some help from Genís in understanding how the code is working, but the complexity of this software really requires a detailed document that explains how and why everything works the way it does. There have been times when I have been unable to help people with CEGMA-related questions because I still can't understand what some of the code is doing.
- Start a FAQ file from day one. This is something that, foolishly, I have only recently started. I could have probably saved myself many hours of email-related support if I had sorted this out earlier.
- Put your code online for others to contribute to. Although GitHub wasn't around when we started CEGMA, I could have put the code up there at some point before today!
- Don't assume that people will use a mailing list for support, or even contact you directly. One thing I did do many years ago, is set up a CEGMA mailing list. However, I'm still surprised that many people just report their CEGMA problems on sites like SEQanswers or BioStars. I probably should have started checking these sites earlier.
- Don't underestimate how much time can be spent supporting software! I probably should have started setting aside a fixed portion of time each week to deal with CEGMA-related issues, rather than trying to tackle things as and when they landed on my doorstep.
- Assume that you will not be the last person to manage a piece of software. There are many things you can do to start good practices very early on, including using email addresses for support which are not tied to a personal account, ensuring that your changes to the code base have meaningful (and helpful) commit messages, and making sure that more than one person has access to wherever the code is going to end up.
In some ways it is very unusual for software to have this type of popularity where people only start using it several years after it is originally developed. But as CEGMA shows, it can happen, and hopefully these notes will serve as a bit of a warning to others who are developing bioinformatics software.